Minimal Spanning Trees¶
[TOC]
Introduction¶
Creating A Tree¶
Trees can be created from the main search page of the database of interest
Stand Alone Trees¶
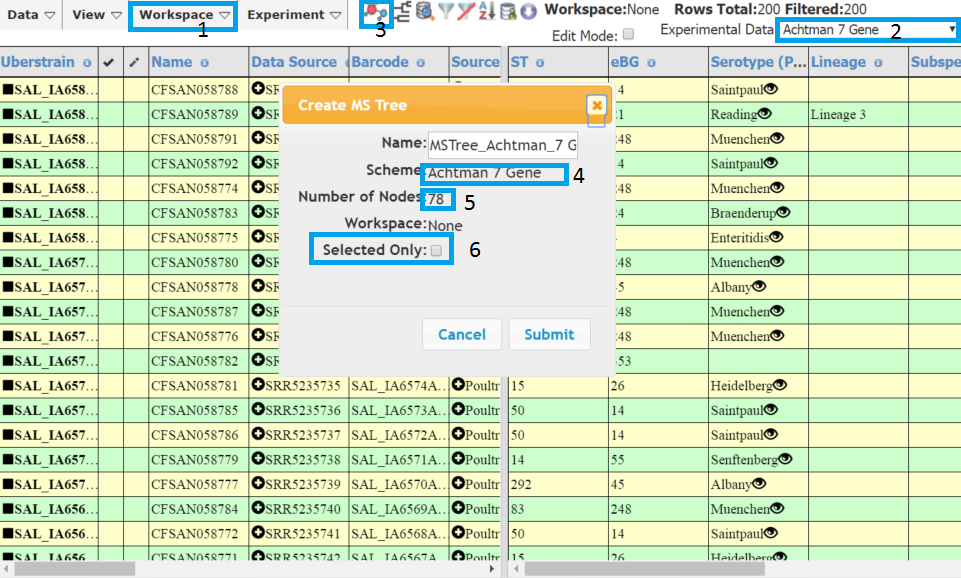
To create a stand alone tree make sure you have all the strains you want to create the in the table. You can copy/paste strains between searches using the Workspace > Copy Selected and Workspace > Paste Strains (1). Next make sure the Experimental data in the right hand panel is the correct MLST scheme (2). Click the MS Tree icon (3) and a dialog will appear reminding you of the scheme (4) and showing the number of nodes (STs) that will be in your tree (5). You will get a warning if your MS Tree contains than 5 nodes. A common mistake is to have 7 gene MLST data displayed that corresponds to only one ST. The tree will only be created from filtered data, so make sure you clear any filters if you want the entire data set. If you just want a tree consisting of a subset of the strains in the current table, you can selected those you want and check ‘selected only’ in the dialog box (6).
After pressing submit, your tree will be sent for computation and another browser window should open. If this does not happen, please ensure that popups are allowed from this site. The tree should appear in the window when the computation is complete. However you can close the window or even close the browser as the tree is computed on a back-end sever. The tree can then be opened by on of the methods detailed below
Tree Connected to A Workspace¶
First of all create a Workspace. Once created an ‘Analysis’ toolbar should appear in the main menu. Choose the ‘Create MS Tree’ under the sub menu items and a dialog box should appear as above. The process is now exactly the same as creating a stand alone tree (follow the instructions as above). The only difference is that the tree will now be connected to the Workspace.
Loading A Tree¶
Stand Alone Trees
- Clicking the load Workspace panel on the top right of the database home page
- On the main search page menu Workspace > load
- Going directly to the tree link e.g. enterobase.warwick.ac.uk/ms_tree?tree_id=3003. If the tree is not public and you are not logged in you will be asked to do so
Trees Connected to a Workspace
- First load the Workspace and then Analysis > load MS Tree or you can click on the tree in the summary dialog
- Go directly to the link e.g. enterobase.warwick.ac.uk/ms_tree?tree_id=3003
Working With Trees¶
When a tree first loads a summary box will appear in which if you have permission you can add a description and any links.Initially the ‘force’ algorithm tries to create the tree, such that all the nodes are spaced out evenly but ensuring the correct lengths between the nodes are maintained. This takes a few seconds and ‘Rendering Tree’ will be displayed in the waiting dialog. One size does not fit all, and thus most trees will require some manual input in order to get a decent looking tree. If you make any changes to the tree, that you want to keep make sure you save the tree using the ‘Save Layout’ button. This ensures that the tree will load quicker as the initial display algorithm does not have to run when the tree is re-loaded. General Manipulation ——————–
- Moving the tree Dragging outside of the tree will move the tree in the direction of the drag.
- Resizing the tree Use the mouse wheel to zoom in or out, with zoom centre, being the current mouse position. You can also zoom in and out using the magnifying glass icons on the left panel.
- Moving nodes Clicking on an individual node will highlight that node and all of its children. All the nodes highlighted then can then be moved in an arc around the connecting parent node.
- Selecting nodes Nodes can be selected by pressing shift and dragging the mouse, all nodes in the rectangle formed will become selected as shown by the red halos. Further selections are added to the current selection and all selections can be cleared by the ‘Clear Selection’ button at the bottom of the left hand panel.
Link Manipulations¶
- Link Length This slider will alter the length of all links. Links are scaled by the number of allele differences between the two connected nodes, thus the relative length of the links will remain constant
- Log scale The links will be scaled by the the log of the number of allele differences. This is useful Where there is a broad range of link lengths
- Hide Links Over All links which correspond to allele difference greater or equal to the number in the text box will become hidden. This is useful for de-cluttering the tree and identifying closely related clusters.
- Max Link Length This will ensure that all link lengths over the value in the text box will remain the same length. Such links will become dashed to indicate they are not accurately scaled. This is useful when you have two or more disparate clusters, but still wanr to show the detail ine each cluster
- Link Labels Whether to hide the link labels, which show the number of allele differences between two nodes
- Font Size This slider controls the font size of the link labels
- Show Tooltip If checked then when you mouse over the links, the value (number of allele differences will be shown)
Node Manipulations¶
- Node Size This slider will alter the size of all nodes
- Relative size The size of nodes which represent more than one strain are log proportional to amount of strains in that node. By using this slider the relative size will change. This is udeful if some nodes representing many strains are too large.
- Node Labels This checkbox signifies whether node labels should be shown
- Font Size This slider alters the font size of the node labels
- Label Text This can either be the ST, strain name or another metadata category. It does not have to be the same category that the nodes are coloured by, however only one value will be displayed, which for nodes representing more than one strain will be the modal (most frequent) value.
- Show Tooltip This checkbox will control whether mouse over a segment will result in the value being displayed in a tootip
- Show Segments By default the nodes are only subdivided into the different values for a category and not each individual strain.If you want this, then check this box. This may make the tree slugish if nodes represent many strains e.g. rMLST trees of entire databases.
Layout¶
By default all nodes are fixed and only move when dragged by the user. However you can unfix either selected or all nodes and they will be move according to the force algorithm. For large trees this will be slow as the all the nodes are constantly updating their positions. When nodes are not fixed, clusters will pull apart and thus the tree may look look less cluttered, however link lengths will no longer be accurate and the visible clusters may be deceptive.
- Unfix Selected Selected nodes will become free and will position themselves according to the force algorithm
- Unfix All All nodes will become free. For large trees, the web page will become less responsive
- Unfix All All nodes will become fixed in their current positions
- Correct Links All link lengths will revert to their correct value i.e. scaled to the number of allele differences between the connected nodes
- Charge This slider alters the charge component of the force algorithm. If any nodes are unfixed, the greater the charge, the more nodes will repel each other (and the less accurate link lengths will become).
- Correct Links This button will ensure all links are scaled to the correct length (the relative number of allele differences between two nodes)
- Refresh This button will recalculate the position of all nodes and redraw the tree
Main Table¶
This section allows cross-talk between the tree and the table of the main search page.
- Load Selected All strains in the selected nodes of the tree will be loaded into the table of main search page. A new window/tab will be opened if one does not exist already
- Filter Selected The table table of main search page will be filtered such that only strains in the selected nodes of the tree will be visible. Only nodes that are already in the table will be filtered. If you want to load new strains, use the ‘Load Selected’ button
- Highlight Selected Any strains selected in the main table will have their corresponding nodes in the tree highlighted with a yellow halo.
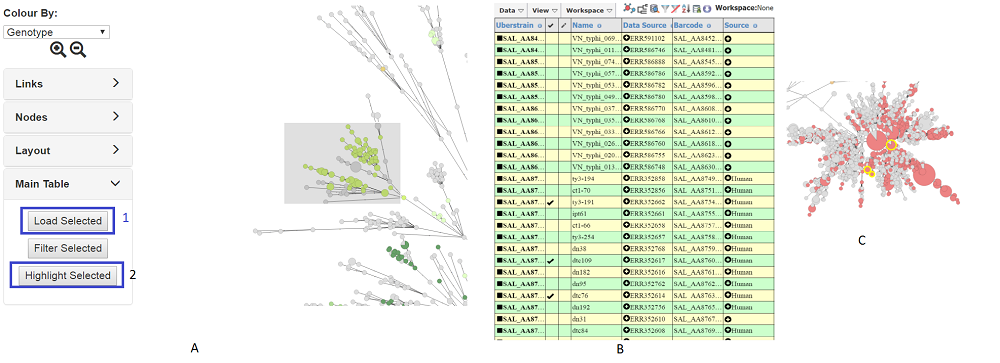
e.g Press shift and drag the mouse to highlight strains nodes in the the tree (A). Highlighted nodes will have a red border. Next press Load Selected (1) in the Main Table tab (if you do not already have main search window open, a new one will appear). All the selected strains will be in the table (B). A link is maintained between the two windows so you can select strains in the main table and press Highlight Selected in the Main Table tab of the tree (2) and the strains will be highlighted with a yellow border (C)
Add Data¶
This section allows you to add data from any scheme associated with the database (including any allele designations) and also to add your own custom metadata
- Experiment Data Choose any experiment/scheme from the dropdown and click on the add icon. All the fields associated with that scheme will then be added to the ‘Colour By’ drop down at the top of the left hand panel.
- Allele Data You can add any locus from the wgMLST (>2000 genes) for the current database. Start by typing the name of the locus or gene e.g. recA and you will get a drop down with suggestions. Click on the one you want and then press the plus icon. Retrieving all the allele calls for large data sets may take a while. When it is ready the tree will be coloured by allele designations (gray nodes show the locus was absent). The locus is also added to the ‘Colour By’ dropdown at the top of the left hand panel.
- Custom Field Custom fields can be added by simply typing in a label name and clicking the plus icon.
- Custom Value You can associate values a custom field by selecting the filed in the dropdown and typing in the name of the value. Then select the nodes you want to give that value to and press the plus icon. The value will be given a default color, but you can change this, see below.
- Upload Values You can upload a tab delimited textfile with the strain barcode linked to the value e.g.
`
Barcode Name My Field
XXX_XX57563 bug a my value
`
You can get a template file by clicking the download icon (or you can save data from the main search table). All other columns apart from Barcode and any custom columns will be ignored, although it may be helpful to have them in the file in order for you to identify strains e.g. Name in this case.
- Adding custom colors Click on the box of the legend item whose colour you want to change and a picker dialog should appear allowing you to choose a new colour.
N.B. Press Save Layout to store all the extra data you have added and any colour changes you have made.
Deleting A Tree¶
- Stand alone Trees can be deleted from the Workspace dialog, which can be accessed either in the database home page ( click the top right panel) or in the main search page (Workspace > Load). Click on the Tree you want to delete and press Delete WS
- Trees can be deleted from Workspace by clicking the edit button in the Workspace Summary dialog. The list of trees should now have a cross by them. Remove Trees by clicking the cross and the pressing Update