Searching strains (Agama example)¶
To illustrate the search function, we will use the Agama dataset described in Zhou et al, bioRxiv 613554.
The browser interface to EnteroBase is based on a workspace concept, implemented as a spreadsheet-like window that can page through 1,000s of entries. However visual scanning of 1,000s of entries is inefficient. EnteroBase therefore offers powerful search functions for identifying sets of isolates that share common phenotypes (metadata) and/or genotypes (experimental data).
Metadata consist of information on the bacterial strain, including isolation year, host and geographical location as well as unique identifier barcodes, strain name, and accession codes that act as hyperlinks to the original data sources. For Salmonella, metadata also includes serovars specified in the SRAs or uploaded by our users. Experimental data includes assembly statistics, and MLST ST (Sequence Type) assignments plus population groupings for all the MLST schemes. For Salmonella, it also includes predictions of serovar using on SISTR1 and SeqSero2. Click here for A full description of metadata fields.
Many entries lack metadata information on serovar, or contain erroneous assignments, and the software predictions are also not fail-proof. We therefore used the Search Strains dialog box to search for entries where the serovar field in metadata contains “Agama” or the software predictions of serovar made either by SISTR1 or SeqSero2 which contain “Agama” (Figure 1). The search results are displayed in two co-ordinated windows, a metadata window at the left and an experimental data window at the right (Figure 2). Figure 2 shows experimental data for cgMLST + HierCC, but other types of experimental data can be chosen from a drop-down list.
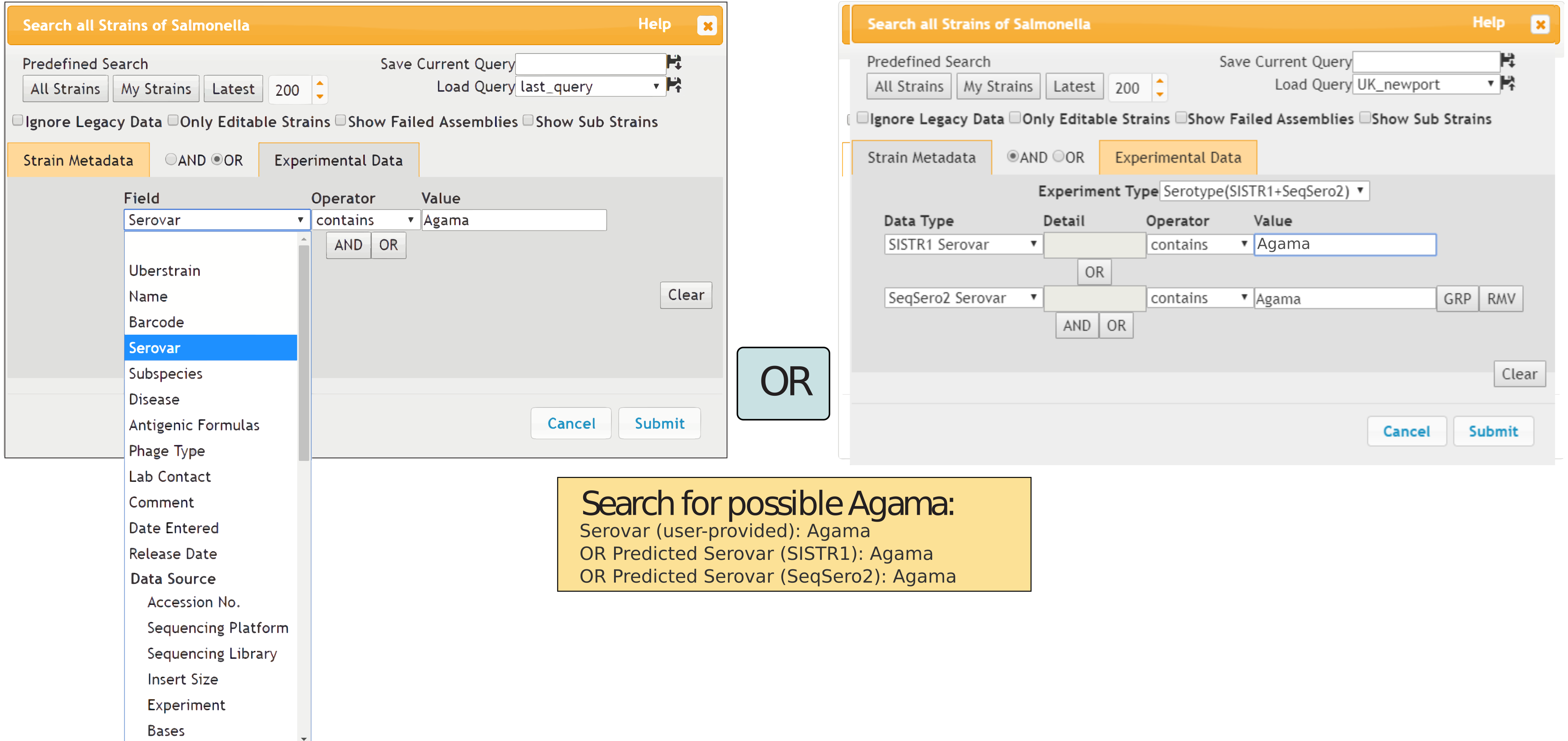
Figure 1: Search strains dialog box Search strains dialog box (above; Figure 1). This dialog box enables searching for all entries that match flexible combinations of metadata (left) and experimental data (right). The dialog box lists the available fields in drop-down lists and supports the operators “contains” (text), “in” (opens a list box for pasting comma- or CRLF-delimited sets of data), “equals”, “>”, “<” and their negations. In this case, the search was designed to identify all Salmonella genomes where the Serovar metadata contained “Agama” or the serovar Agama had been predicted from the genomic assembly by SISTR1 or SeqSero2.
All entries that matched the search are shown in a spread-sheet like interface consisting of a metadata window (Figure 2; left) and an experimental data window (Figure 2; right), which was sorted in ascending order by clicking on the column header for HC2000 at the right.
The drop-down box at the upper right for Salmonella allows a choice between experimental data: 7-gene MLST, Assembly statistics, Annotation downloads (General Feature Format [GFF] or GenBank format [GBK]), wgMLST (Alleles and ST numbers), Serotype prediction, rMLST (Alleles, ST numbers, reBG, serovar prediction), cgMLST V2 and HierCC (Alleles, ST numbers and HC clusters based on cgMLST at 13 levels ranging from HC0 to HC2850).
Experimental Data also includes Custom Views and User Defined Fields. Similar experimental data are available for each of the taxa covered by EnteroBase. If the experimental data include ST designations, clicking on the GrapeTree symbol in the menu will create a GrapeTree of allelic differences. The red box highlights that multiple Agama entries all share the HC2000_299 HC cluster.
Right clicking on any cell within the right window gives access to a List Box offering Select All (entries in browser; indicated in second column from left), Unselect All, Find STs (with up to a user-defined number of different alleles). Get at this level (find all Strains with the same HC cluster designation or eBG/ST Complex), Download Allelic Profile (all entries in the spread-sheet) and Download Allelic Profile (Selected Only).
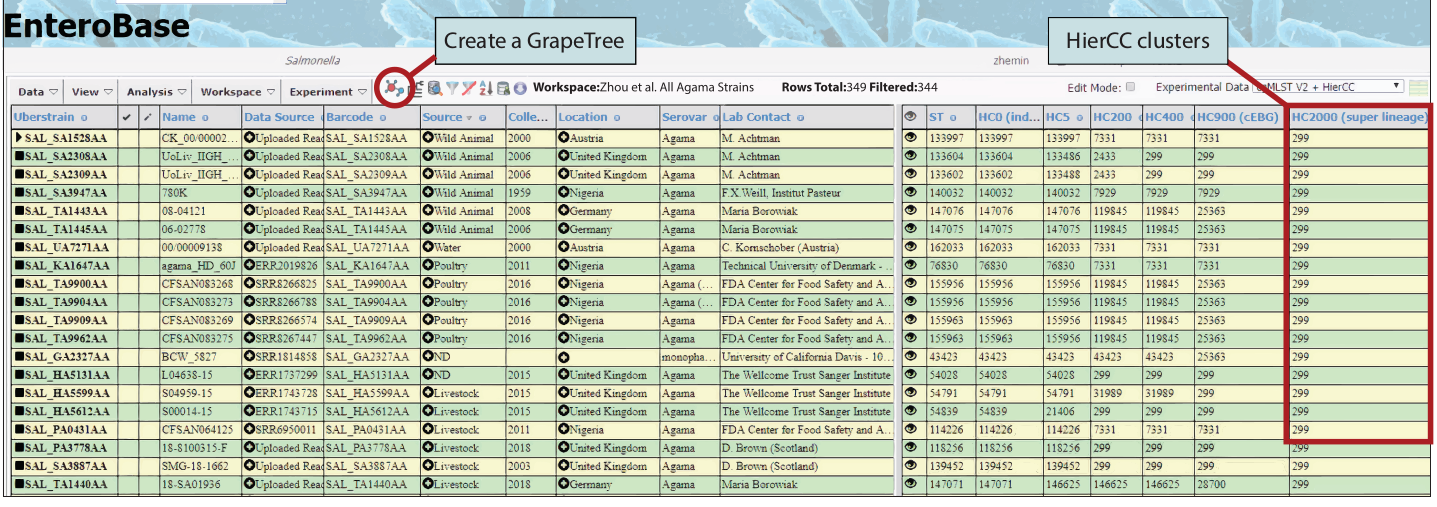
Figure 2: Results of the search