UserSpace OS¶
In this example, we will walk through the iterative process of exploring data around strains of interest, and sharing this information with collaborators.
In our publication Zhou et al, bioRxiv 613554, Our initial phylogenetic analyses indicated that Salmonella enterica ser. Agama consisted of multiple micro-clusters, one of which included all Agama isolates from badgers. However, these results were possibly distorted by a highly skewed geographical sampling bias because almost all isolates were from England. We therefore formed the Agama Study Group, consisting of our colleagues at national microbiological reference laboratories in England, Scotland, Ireland, France, Germany and Austria. The participants were declared as buddies within EnteroBase in order to provide shared access to the Workspaces and phylogenetic trees in the ‘Zhou et al. Agama’ folder (Figure 1; left). That folder has now been made publicly available and can be found at https://enterobase.warwick.ac.uk/species/index/senterica.
Analyses can benefit from aggregating fields from multiple sources of experimental data and/or User-defined Fields for text annotations in a user-defined Custom View. A new Custom View was established for the Agama Study Group (Figure 1; right), saved in the Agama folder, and thereby also shared with the entire Study Group. This view thereby remained private during the initial analyses, and became public when the Agama folder became publicly visible. Members of the Agama Study Group were requested to sequence genomes from all the Agama strains in their collections, and to upload those short reads to EnteroBase, or alternatively to send the bacterial strains or their DNAs to University of Warwick for sequencing and uploading. The new entries were added to the ‘Zhou et al., All Agama Strains’ workspace (Figure 1), which now contains 344 genomes.
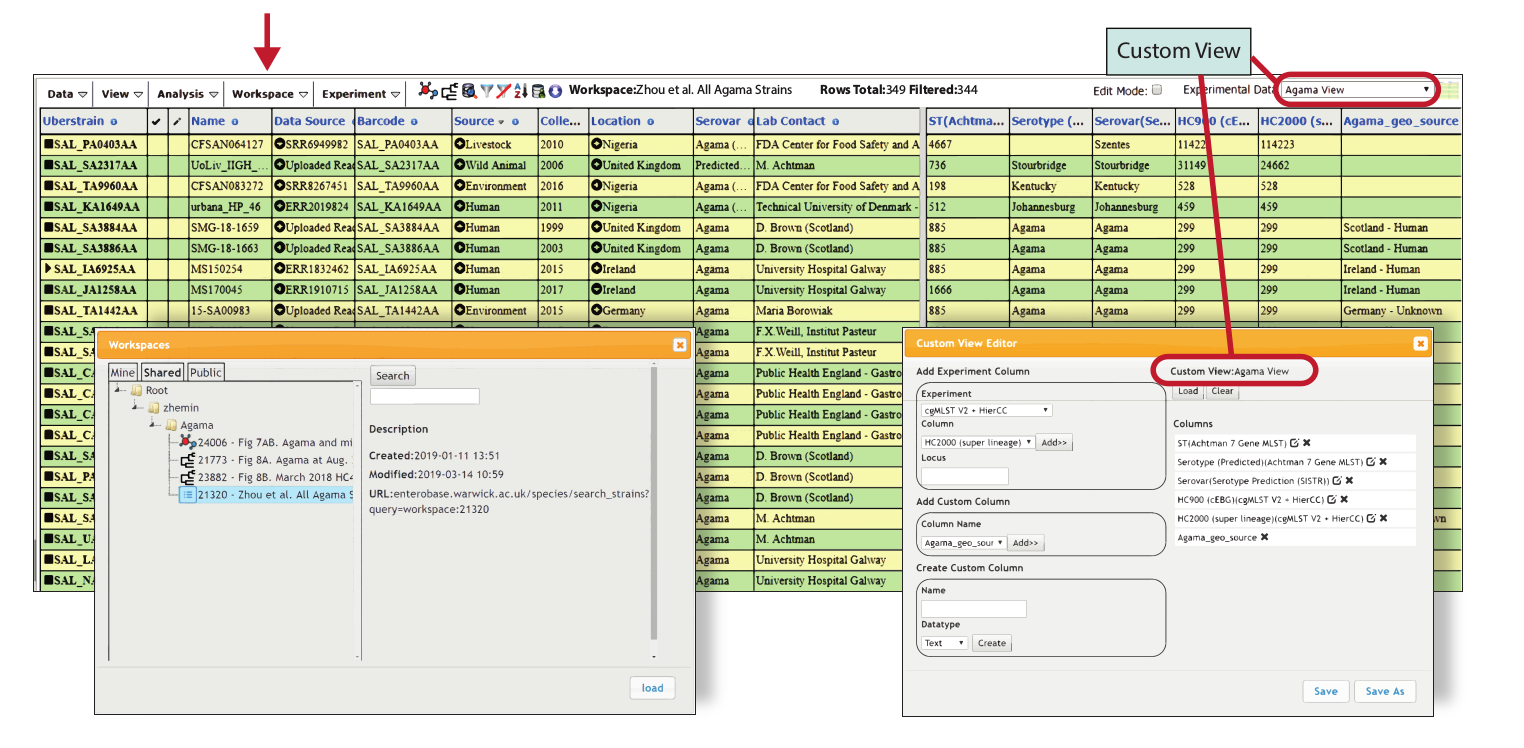
Figure 1: UserSpace OS
The menu item labelled Workspace (red arrow) allows access to multiple tools implemented within the UserSpace OS for working with workspaces, including Empty (contents). Save (current changes), Summary (Description and external hyperlinks), Save As, Load, Copy Selected (entries), Paste Strains (which were copied in a separate workspace), Add Selected to Workspace (selected in current workspace to a different workspace), Delete Selected (from current workspace). “Load” opens the dialog box shown on the left, which provides an overview of the user’s personal area (Mine), of workspaces shared by other users (Shared) and of publicly available workspaces (Public). The experimental data tab at the upper right shows that the Custom View “Agama View” has been chosen whereas the dialog box shows how this view was created within Custom ViewNew/Edit. Custom views can consist of any combination of Experimental Data fields and User Defined Fields (text). They can be saved in the user’s WorkSpace folder, and shared with other users in My Buddies (left menu).