Adding updated Serovar Information¶
The input data consists of an Excel file containing 3 columns Name, Serovar and Antigenic Formulas.
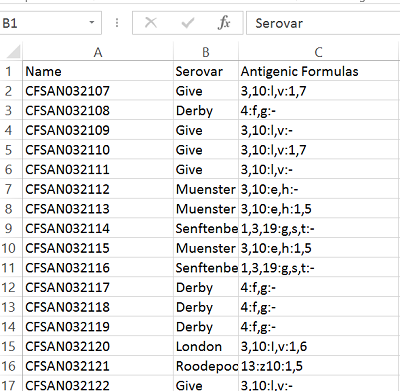
Formatting the Input Data¶
You cannot use the strain name as the key to upload the modified metadata, as it is not unique. Instead you have to use Accession or Barcode. In this case we are going to use Barcode and in order to retrieve this from EnteroBase, you will first have to load the strains into the main search page.
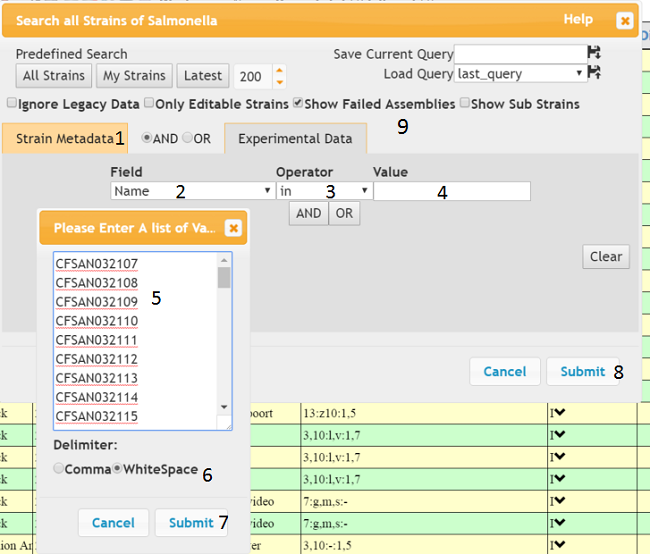
In Excel, select and copy the names of the strains. Then in the ‘Strain Metadata’ tab (1) of the query dialog, select ‘Name’ from the ‘Field’ dropdown and ‘in’ from the Operator dropdown (3). When you click on the ‘Value’ input (4) another dialog will appear and you can paste the strain names from excel into the dialog’s text box. Make sure the delimiter is WhiteSpace (6), then press ‘Submit’ on the sub-dialog (7) and the ‘Submit’ on the main dialog (8), The strains should then appear in the main table, make sure you have the ‘Show Failed Assemblies’ checkbox checked if some of the strains you queried have no valid assembly.
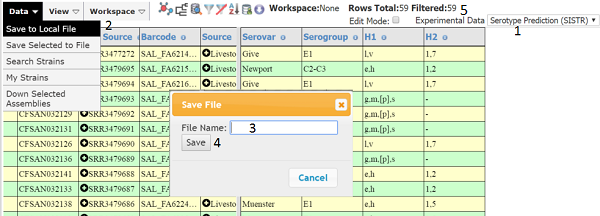
Make sure the number of strains is the expected value (5). Then save to a local file, which is in the ‘Data’ section of the main menu (2) . Type an appropriate name for the file (3) and press ‘Save’ (4). The file will be saved to the download folder used by your browser as tab delimited text. In this case, the ‘Experimental Data’ was changed to ‘Serotype Prediction’ (1) because the edited metadata contained Serovar and thus this enables a sanity check of the edited data. You can then open the file in excel and to make it cleaner you can delete all columns except Name and Barcode. Next, you need to associate your edited values with the Barcode. This can be done with the VLOOKUP function, or simply sorting both tables on name and then cut and pasting the Barcode column to the table containing the modified Serovar and Antigenic Formulas (see below).
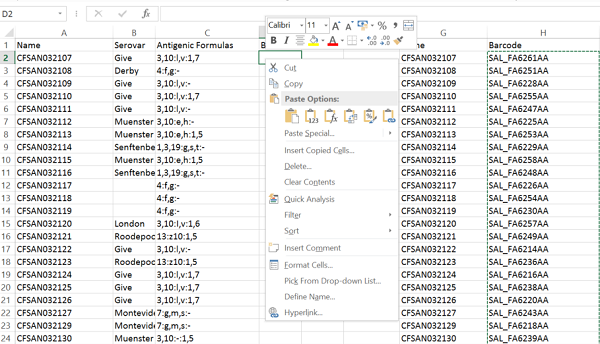
You can then save this table as tab delimited text and upload it to EnteroBase.
Uploading the File to EnteroBase¶
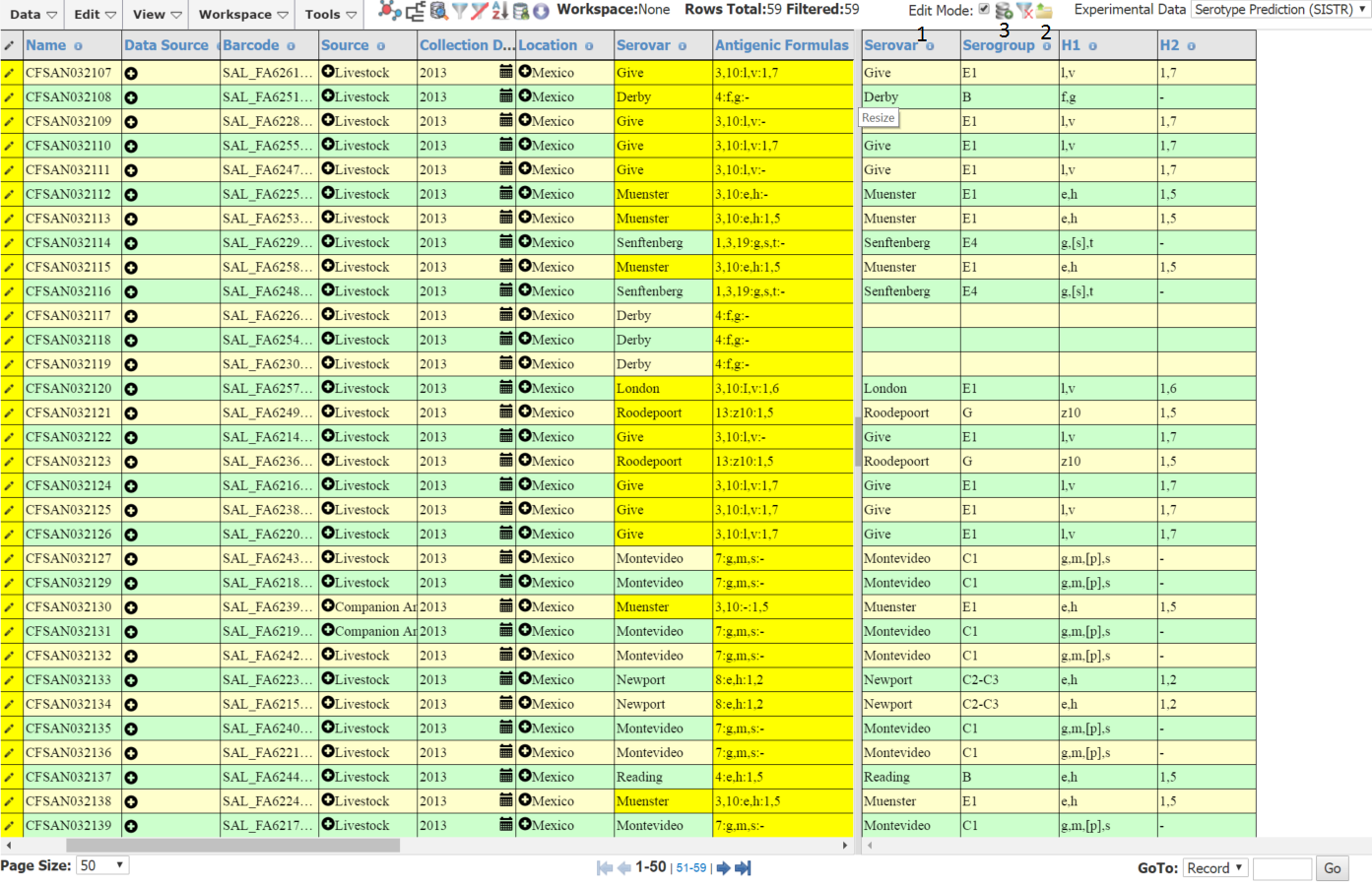
To upload the edited file, make sure you are in edit mode (1), then click the upload file icon (2). The strains should appear in the table and any modified cells should be shown in yellow . Check the changes and make any extra directly in the table. When you are finished click the update database icon (3). You should get a dialog telling you that the records have been updated successfully.