Searching EnteroBase¶
By clicking through Search Strains you are presented with the Search menu with the data panels in the background. Once you submit a search, these panels will fill with your search results.
Let’s perform a search together by following these steps:
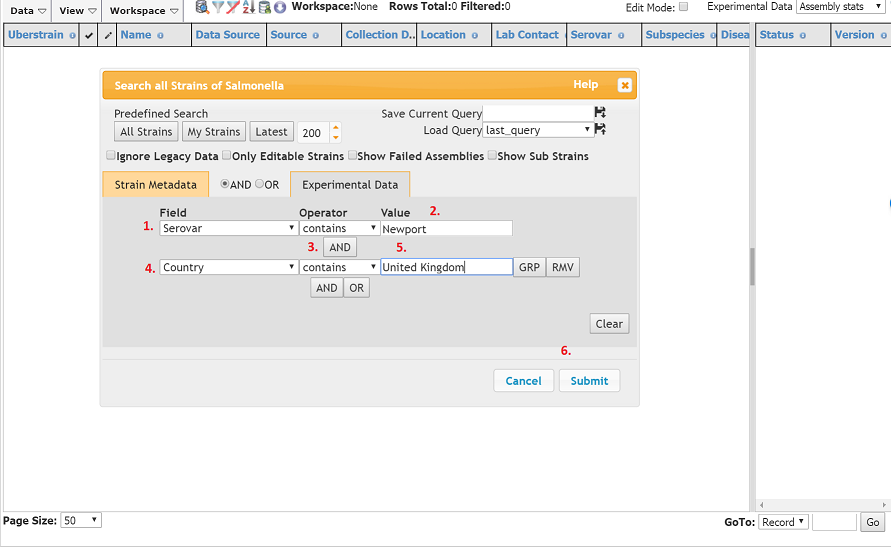
- Click the Field dropdown and select Serovar. N.B. A Serovar is just a sub-division of the species based on antigenicity, in Salmonella these are often named after geographic locations.
- In the Value field to the right, type in Newport (a common Salmonella serovar). You should notice that a dropdown will appear below giving suggestions. You can click on ‘Newport’ in this list or continue typing the full word.
- Click the AND button. We want add an additional condition to our search.
- Click the 2nd ‘Field’ dropdown and select ‘County.
- In the ‘Value’ field to the right, type in ‘United Kingdom’. You should notice that a dropdown will appear below giving suggestions. You can click on ‘United Kingdom’ in this list or continue typing the full phrase.
You have now prepared a reasonably complicated query. We are searching for strains of serovar Newport that were isolated in the United Kingdom. Click ‘Submit’ (6).
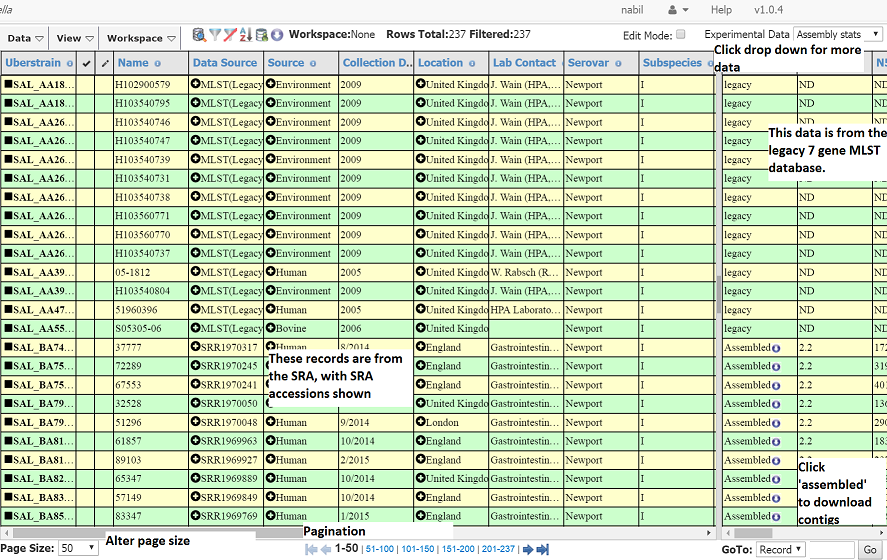
The exact data will change as new data would have been added since the preparation of this page. However, you should see a number of strain records with metadata on the left pane and experimental data on the right pane.
The first few rows show records from the legacy MLST database (http://mlst.warwick.ac.uk), and as such they have ‘MLST(legacy)’ as the data source. These data are derived from Sanger Traces and have no NGS data, so a number of assembly statistics and genotyping information is blank. While EnteroBase shows past MLST data, EnteroBase does not accept new data based on Sanger Traces.
The other rows (at the bottom) are derived from sequenced reads from the SRA. Their status shows they’ve been assembled and the data Source shows the SRA accession number.
Let’s revisit the search function. It may be blank, which means repeat the steps from before to search for strains of serovar Newport from the United Kingdom.
## Saving and loading queries As you can see, it is a little time consuming to repeatedly enter all the information for complex queries. EnteroBase has a feature where you can save an important query and load it on demand.
To save the current query for later:
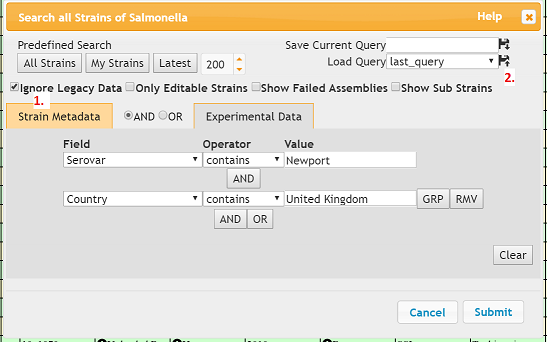
- Enter an informative query name e.g. ‘Newport_UK’ in the text box right of ‘Save Current Query’ (2).
- Click the ‘Save’ button (floppy disk icon with a down arrow).
Now press the ‘Clear’ button near the bottom right and try loading your query
To load a query:
- Click the dropdown, and you should see your query name e.g. ‘Newport_UK’. Select it.
- Click the ‘Load button (floppy disk icon with an up arrow).
Advanced query functions¶
There are a number of extra options to enhance your searches:
- ‘Ignore Legacy Data’: You can hide legacy data by checking this box (1)
- ‘Only Editable Strains’: You can show strains only you own/or can edit. Usually there will be no results if you haven’t uploaded anything to EnteroBase.
- ‘Show failed assemblies’: Show assemblies that have failed the quality control. These are usually hidden. These strains will not have any genotyping or other analysis run on them, but it maybe useful to check the assembly statistics and download the contigs to see what went wrong.
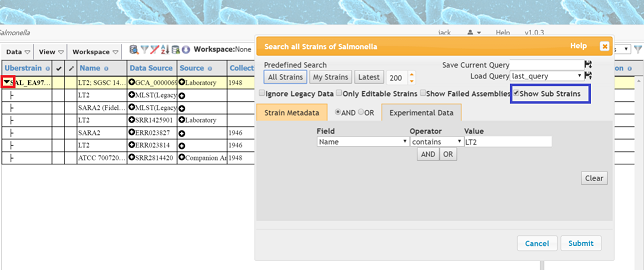
- ‘Show sub strains’: Some strains have been grouped together for various reasons (see Section Uberstrains). These are usually hidden from search results but are shown if this is checked.
There are also predefine searches that can be run with one-click, under ‘Predefined Search’ in the top right:
- ‘All Strains’: Fetches all strains records for the whole database. This can be slow on large databases.
- ‘My Strains’: Fetches strains that belong to you.
- ‘Latest XXX’: By setting a number in the number field, the search will fetch the last X number of strain records entered in the database.
Uberstrains¶
Most bacterial isolates/strains in EnteroBase are linked to one set of metadata and one set of genotyping data. However, some entries have two or more sets of genotyping data. For example, EnteroBase includes some strains for which legacy MLST data from classical Sanger sequencing exists in addition to MLST genotypes from genomic assemblies. Similarly, some users have uploaded the same reads to both EnteroBase and SRAs, and both sets of data are present in EnteroBase because it automatically imports all new SRA records. In still other cases, genomes of the same strain have been sequenced by independent laboratories, or multiple laboratory variants have been sequenced that are essentially indistinguishable (e.g. S. enterica LT2 or E. coli K-12).
You may have noticed the Uberstrain column at the very left of the strains table and wondered what it is all about. Certain records are duplicated in that there are many entries for what is essentially the same strain. This can skew analysis because your analysis may produce false clusters, which in reality are just the same strain. Thus in Enterobase such entries are merged and a single Uberstrain is created. Normally only the Uberstrain is shown and so you do not need to worry about this de-replication. However you can still examine the sub-strains associated with an Uberstrain (see below)
- Go the Salmonella database and search for LT2, make sure that the Show Sub Strains box in checked in the query dialog (blue box). A single record should load but it will have an expand icon in the Uberstrain column (red box). Clicking on this icon will show all the sub strains associated with this master strain. The master strain is usually the most complete (in this case it is the complete closed genome).
Downloading Data¶
The current data can always be saved to file by `Data -> Save To Local File`.
Because of browser restrictions, the data is actually treated as a download so
it may probably end up in the downloads directory that your browser uses. Some
browsers however let you choose the location. The file is just a tab delimited
text file, which can be opened in any spread sheet. The file will contain all
the strain metadata and any associated experimental data in the right hand pane.
For large schemes, this data is not very useful therefore Enterobase enables you
to download all the allele information separately.
In the Salmonella database query on serovar equals Dublin. By default the experimental data will be assembly stats. In the Experimental Data dropdown choose cgMLST (3020). The data in the right hand panel with then show the cgMLST ST for each record, which is not very useful (but then would you want to look a 3020 columns of allele numbers). Right Click on the right hand panel and select Save all. A dialog will appear showing the progress of retrieving the information (100 records are obtained at one time). Once the data has been retrieved, it can be saved as with any other file (type a file name in the text box and press save).
Editing Metadata¶
- Load all the records in the test database and click on the Edit mode check box.
You will get a dialog with some information – click OK. Records with a pencil icon (blue box) show that you have permission to edit the metadata (In this case, it will only be the four strain you have uploaded). Click on any cell and alter its content and the cell should turn yellow. Once you have made all the changes you require you need to upload these changes to the database (press the upload changes icon - red box) or right click on row containing an edited cell and select Upload Changes in Row.
After updating changes the cells should turn back to their normal colour and a
dialog will inform you whether the update has been successful.
A Search and Replace (`Edit -> Search and Replace`) and undo function (`Edit -> Undo` or ctrl+Z) are
available but for large scale editing it may be easier to load the data into Excel.
- Click on the My strains Icon or
`Data -> My strains`or and the four strains you uploaded previously should be present – all of which you have editing permission. Then save them to a local file`Data -> Save to Local File`. Open this file in excel and change some values. Then re-save the file (making sure it is in tab delimited text). Reload the file into Enterobase, by first clicking on the edit mode check box and then clicking The Load Modified File icon (purple box). Any changes or errors in your modified file will be shown as yellow or red cells.
In edit mode you can send jobs / assemblies on any strains that you have editing
rights to by using `Tools -> Assemble Selected` or `Tools -> Call Scheme for Selected`.