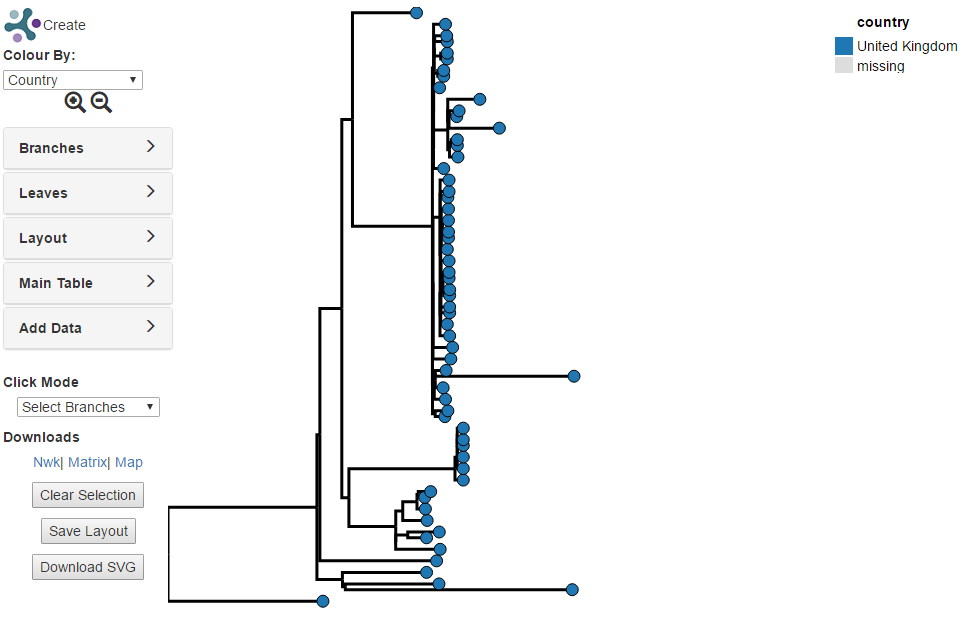
Creating a SNP Tree¶
In this tutorial a tree of all predicted Salmonella typhi for samples from
London will be created. First, the initial search will be conducted in the
Salmonella database. Either click on the link to “Search Strains” on the
Salmonella home page at
https://enterobase.warwick.ac.uk/species/index/senterica (or if you are
already on the Salmonella related pages select “Search Strains” from the
Tasks menu i.e. Tasks -> Search Strains). This will open a dialogue box
to set up a new search and will initially be on the “Strain Metadata” tab.
Select “City” (under heading “Location”)(1) for “Field” and “London” for
“Value”(2).
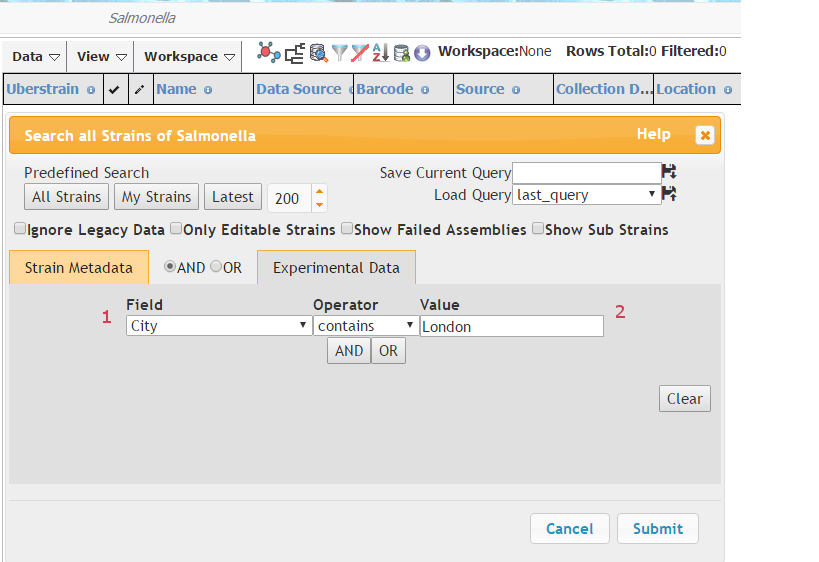
Then go to the “Experimental Data” tab (1 in image below), and select “Serotype Prediction (SISTR)” (2) from the Experiment Type dropdown menu. Select “Serovar” from the “Data Type” dropdown menu (3), equals from the “Operator” dropdown menu (4) and type “Typhi” in the “Value” text box (5) Press submit and a “Processing Query” box should appear.
Tree Construction¶
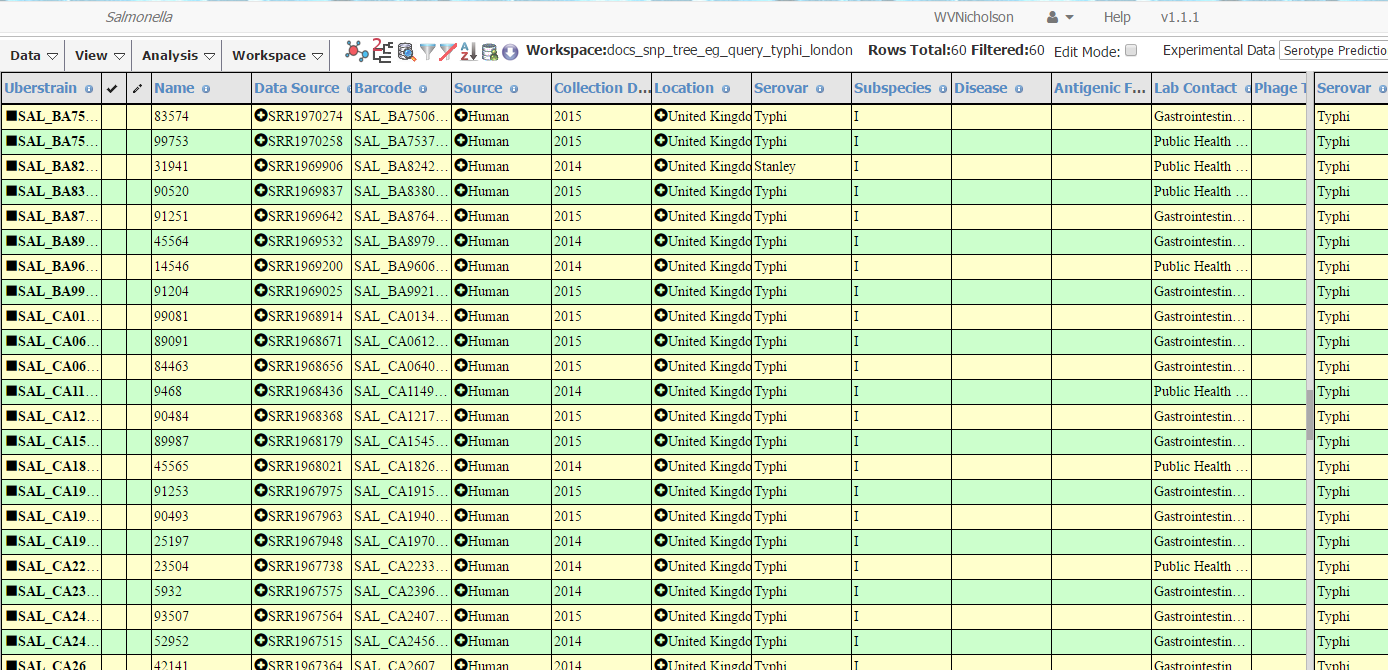
After a few seconds, the strains searched for should appear in the table.
After the query, the number of strains will appear in the top bar (1 in image
below). (This number may differ from the image below due to new strains in
the database.) Then in order to compute a SNP tree click on the tree icon
(with a tree made up of straight lines) on the top bar (2). (Mousing over the
icon will display the message/ tool tip “Create SNP Project” in most modern
web browsers.) A dialog box “Call SNPs on strains” will appear. Enter a
suitable name for the SNP project (making a note of the chosen name) and
choose a reference. (For the purposes of the tutorial using the default
reference is sufficient.) Then press the “Submit” button. A dialog box
“Warning: Your SNP job has been successfully submitted” should then appear.
SNP tree related pipelines will run in the order [refMasker], [refMapper],
[refMapper_matrix] and [matrix_phylogeny] as described [in the section on SNP
trees on the page about the EnteroBase
backend](EnteroBase%20Backend%20Pipeline#markdown-header-snp-trees).
Depending on the size of the job, these pipelines may take a long-ish time to
run. The individual jobs running the pipelines can be monitored by going to
Tasks -> Show My Jobs as described [on this page](Jobs).
Viewing the results¶
Once all of the SNP pipelines have finished then the results can be examined
by going into Workspace -> Load Workspace. (Since the SNP pipelines may
take a long time to run in the case of a large job, you may have to re-visit
the database search page first, in order to have access to the menu that
allows you to load the results from the completed SNP project job.) Then
select the entity with the earlier chosen name for the SNP project and press
the “load” button. Then 2 independent windows will open - a window displaying
the tree and also a JBrowse window. The window displaying the tree will also
have a dialogue displayed internally which may optionally contain a
description and links. Press the “OK” button.
The page on [SNP Projects](SNP Projects) describes options for displaying and manipulating the trees.