Find similar STs¶
A common question with a given strain is ‘Which other known strains are closely related it?’ This is easy to answer with EnteroBase using the ‘Find Similar STs’ option.
Most strains in EnteroBase have an ST and allele profile designated for each of the genomes in EnteroBase. You can actually search for similarity between allele profiles. This currently applies for cgMLST, MLST, and rMLST.
Here is a worked example, I am interested in this strains, ‘2016K-0033’ and I want to retrieve all similar genomes for deeper SNP analysis.
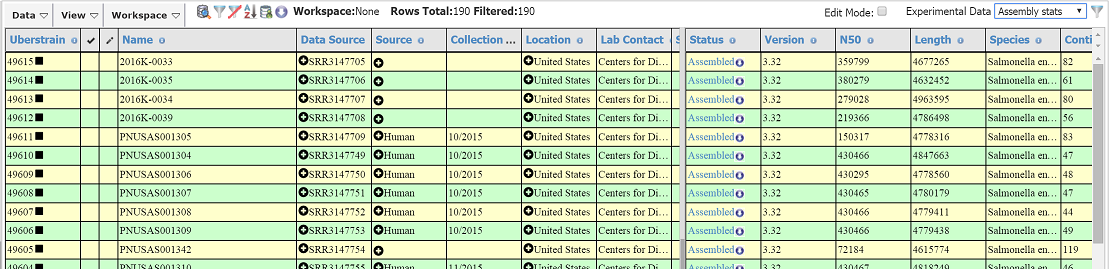
- I want to find similar strains based on rMLST, which is (usually) more discriminant than classical MLST.
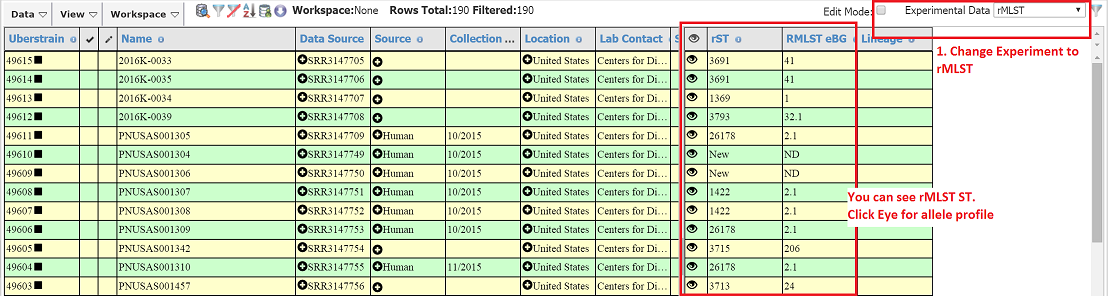
- Right click to find similar strains
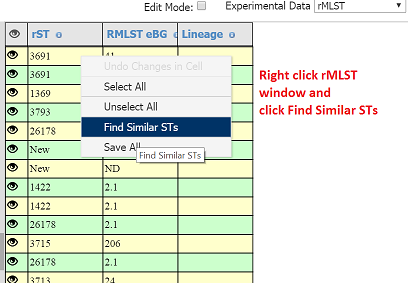
- You can choose how similar other matches should be, in this case I set 52 out out of a total of 53.
- **These are our results! There are 199 similar strains, within 1 rMLST allele difference. We can immediately see that our strain of interest is an Oranienburg (that’s what all its cousins are). **
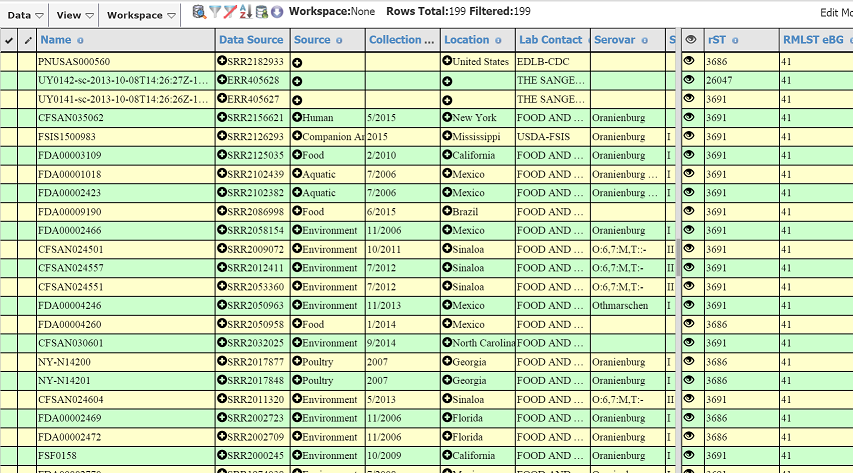
- **Select a bunch and download the assemblies and off we go. **
