Tutorial 1B: Basic Usage of GrapeTree (Stand-alone)¶
The procedure for working with GrapeTree in EnteroBase and stand-alone are identical, once you have loaded your data. This tutorial assumes you are using the stand-alone version. There are other tutorials specifically for the EnteroBase version.
You will need to install GrapeTree, Installing stand-alone GrapeTree.
About this dataset¶
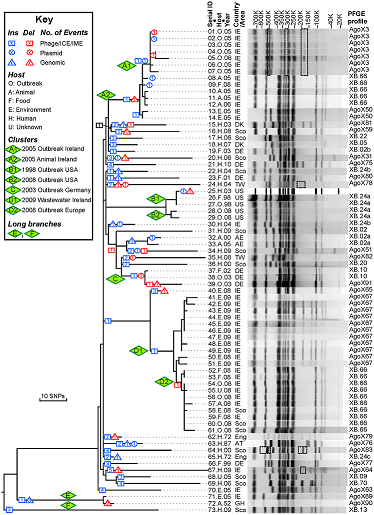
To learn the basic usage of GrapeTree we will be using data presented in Zhou et al. “Neutral genomic microevolution of a recently emerged pathogen, Salmonella enterica serovar Agona”. PLoS genetics 9.4 (2013): e1003471. We will try to replicate the tree presented in Figure 1, which shows phylogeny of serovar Agona including a number of outbreaks (green diamonds) across the world.
1. Loading data into GrapeTree with the stand-alone version¶
Download the sample data as shown below.
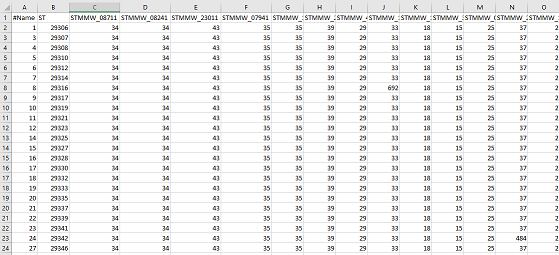
The profile file includes *Salmonella* cgMLST data from EnteroBase for the strains described in the paper. The first row contains the headers for each column “Name”, “ST” (Sequence Type), and each subsequent column are the names of locus within the cgMLST scheme.
If you wish to use your own profile:
- Profiles must be tab or comma delimited.
- You MUST include a “#” symbol as the start of the row. Note in the example below that “Name” is in fact “#Name”.
- You may use a SNP matrix, which is the same format as the Agona example, with single nucleotides (A,T,G or C) substituting for the numbers.
The metadata file is slightly modified version of what is available in EnteroBase.
If you wish to use your own metadata:
- Metadata must be tab or comma delimited.
- One column must be labelled ID and these values should correspond to the names in the profile file.
2. Install and Start GrapeTree¶
Start GrapeTree as described here, Installing stand-alone GrapeTree.
3. Load in the Profile file and Metadata¶
You should now see the GrapeTree interface with the splash screen. You can either drag-and-drop the profile file into the window or click Load Files and navigate to the file through the file browser.
You will be prompted to select the Parameters For Tree Creation. The method should be set to MSTTreeV2 in the dropdown and then click OK.
Repeat the process with the metadata file. Either drag-and-drop the file into the window or click Load Files and navigate to the file. You should now see the tree colored with a metadata field as shown below.
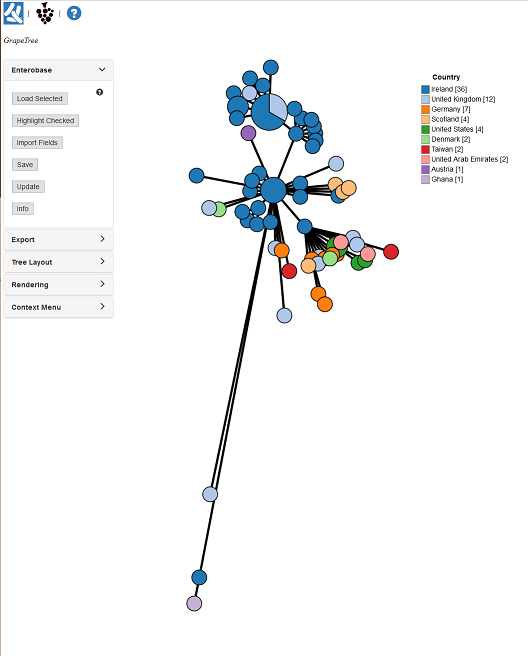
2. Basic Orientation¶
Let’s stop and orientate ourselves with the GrapeTree interface:
- These are links to important webpages (Top left). The Left icon opens a new browser page to EnteroBase, while the right opens the GrapeTree GitHub page.
- The text here (Top left) shows may show the filename of the file we just loaded in.
- This set of panels (Left) contains all the options for customizing our tree. Currently the Input/output panel is open and gives options to load trees/profiles, load metadata, and options to save our work.
- The GrapeTree Tree itself (Centre), the figure is interactive. Each circle is a Node and each line is a branch. Node size is dependent on the number of strains within that node. Branch length varies on the distance between nodes.
- The Key/Legend (Right) for the colour coding. You can change some settings by right clicking on it.
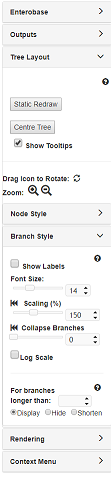
6. Modifying the Tree Layout¶
The Tree Layout panel allows global changes to tree layout, nodes and branches and has some important navigation features. Try playing around with each of the settings to see what they do. Generally:
- You can drag the sliders to change the value.
- You can also directly modify the value by clicking on the value box, typing in a new value, and pressing enter or clicking out of the box.
- Click the refresh icon (the rewind icon) to reset the value to default.
Specifically under Tree Layout > Branch Style:
- Scaling: will uniformly increase the scaling for all branches. For instance, setting it to 200% will double the length of all branches relative to the default setting (100%); whereas 50% would halve it.
- Collapse Branches: will collapse all branches under a certain length. The length value shown is the real branch length for the tree. To see the lengths for all branches, check the Branch Labels under Branch Style. The slider is scaled relatively, so moving it all the way to the right will collapse all the nodes giving you a pie graph.
- Log Scale: if this is checked (has a tick in the box), all branches will be Log-scaled. This is useful for trees with a wide variety of branch lengths.
Specifically under Tree Layout > Node Style:
- Node Size: will uniformly increase the scaling for all Nodes. For instance, setting it to 200% will double the size of all Nodes relative to the default setting (100%); whereas 50% would halve it.
- Node Scaling: This will exaggerate differences in node size. In the case of the of the Agona dataset, all nodes include only one strain so there will be no effect.
For this tutorial set the Branch length to 150% (as shown below) and we shall continue.
7. Styling the Branches¶
Under Branch Style we can also modify the look of the Branches in our Tree. We can show branch labels by checking the Branch Labels option and change the font size with the Font Size slider or by entering a new value in the box. If the Mouseover info box is checked we can see the branch length when we have the mouse cursor over a particular branch.
The tree of the Agona dataset has very long branches. Enter 100 in the box next to For branches longer than and set it to ‘Shorten’. This will shorten the branch length and change the line to be dashed, which indicates the branch length is not to scale.
8. Node settings¶
Under Tree Layout > Node Style we can modify the look of the Nodes in our Tree. We can show nodes labels by checking the Node Labels option and change the font size with the Font Size slider or by entering a new value in the box. If the Mouseover info box is checked we can see details for that node when we mouse over.
We can also set the colour coding of the Nodes. For the Agona dataset, Set Colour by to “Agona_Clusters” to show the outbreaks as defined the original paper. See if you can your figure to look like mine.
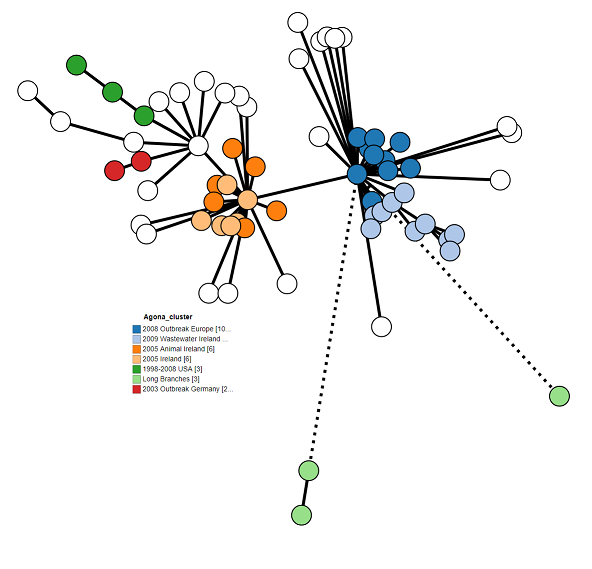
9. Final modifications¶
The tree is looking pretty good, but we can make it a bit clearer. Try playing around with all of the different options to come up with the best looking tree. Here’s what I came up with:
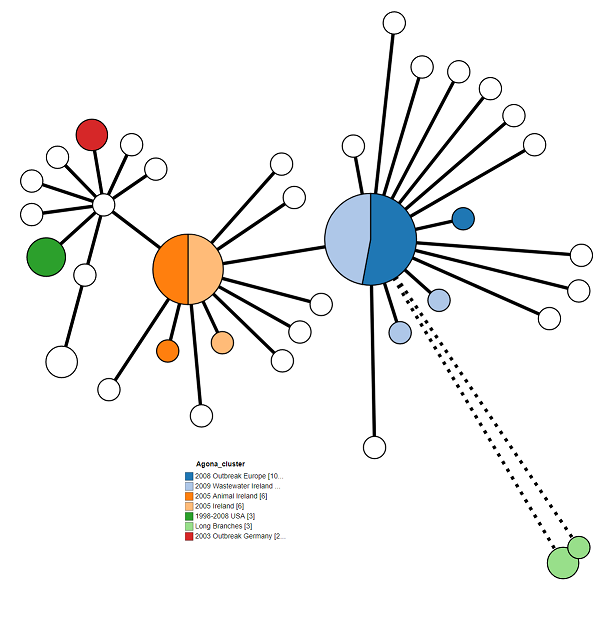
These are my settings:
- Tree Layout > Branch Length: 150%
- Tree Layout > Collapse Branches: 10
- Branch Style > Shorten branches longer than: 100
- Node Style > Colour by: Clusters
- Node Style > Node Labels: Unchecked/Off
10. Exporting our work¶
Your tree can be save in either GrapeTree’s JSON format, as a Newick tree that can be loaded into other phylogeny programs and as a Scalar Vector Graphic (SVG), which is an image format that you can edit in publishing software like Inkscape or Adobe Illustrator. If you would like a raster image (JPEG, PNG, BMP) of the Tree, just use the screenshot feature of your computer.