Downloading schemes from the website¶
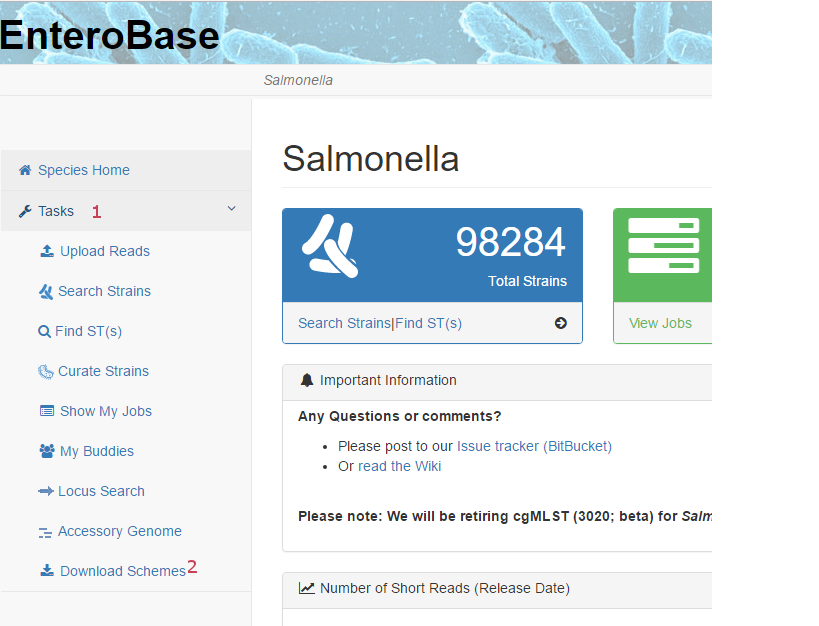
In order to download schemes from the website for a particular database, go to the home page for that database
(e.g. for the Salmonella database)
and select Download Schemes from the Tasks menu (2 in the image below of the top
left part of the database home page) on the left hand side
(i.e. use Tasks -> Download Schemes). This may require clicking on “Tasks” (1) in order to open up the
Tasks menu.
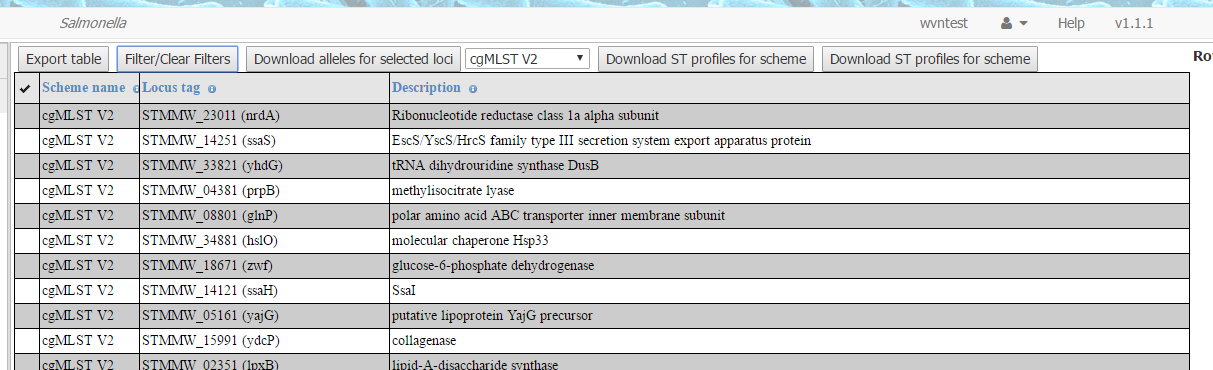
This then navigates to the page to download schemes for the current
database (part of the top half of this page for the Salmonella database is shown below as an example). (It is also possible to get to this page
from other pages, such as the page for searching strains, by using Tasks -> Download Schemes, providing that they have the Tasks menu available on the left hand side of the page.)
Downloading ST profiles for scheme¶
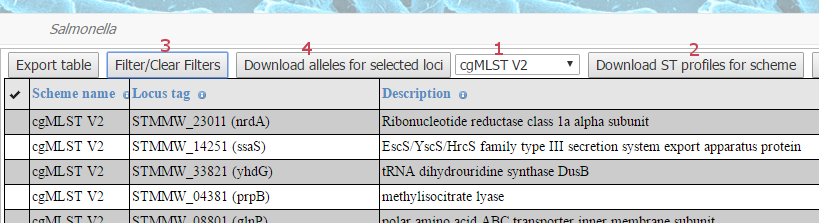
Then, in order to download a scheme, select from the dropdown menu (1 in the image of the buttons below) with a
choice of genotyping schemes available to download from EnteroBase (currently wgMLST, cgMLST V2 and Achtman 7 Gene
in the case of Salmonella), and press the button Download ST profiles for scheme (2) just to the right of the dropdown menu - which will download a gzipped text file containing the allele numbers for each ST (for all of the STs currently in the database).
Downloading alleles for selected scheme¶
Pressing the button Filter/Clear Filters (3) opens a Filter Data dialogue. Changing the scheme in this
dialogue and pressing the Filter button (in the dialogue box) will then show the loci for that scheme.
Then select individual loci by clicking in the column headed with a tick icon in the boxes next to the desired loci
and press the Download alleles for selected loci button (4). This will download a gzipped FASTA file for
each selected locus which will contain the allele sequence for every allele ID currently in the database for that
locus (with the sequence name in the file obtained by combining the locus name with the allele ID, delimited by an
underscore character). For example, doing this with all of the loci in the Achtman 7 Gene scheme will download 7
files.
These files can also be accessed from the directory https://enterobase.warwick.ac.uk/schemes/