Getting started with Enterobase API¶
All API activity needs to requested through HTTP Basic Authentication and authenticated with a valid token. You must have an account on the EnteroBase website to get your Token.
If you have API access to a database¶
Your API token should be displayed under ‘Important information’ in the main database dashboard. (To access a database dashboard, from the main page of https://enterobase.warwick.ac.uk, click ‘Database Home’)
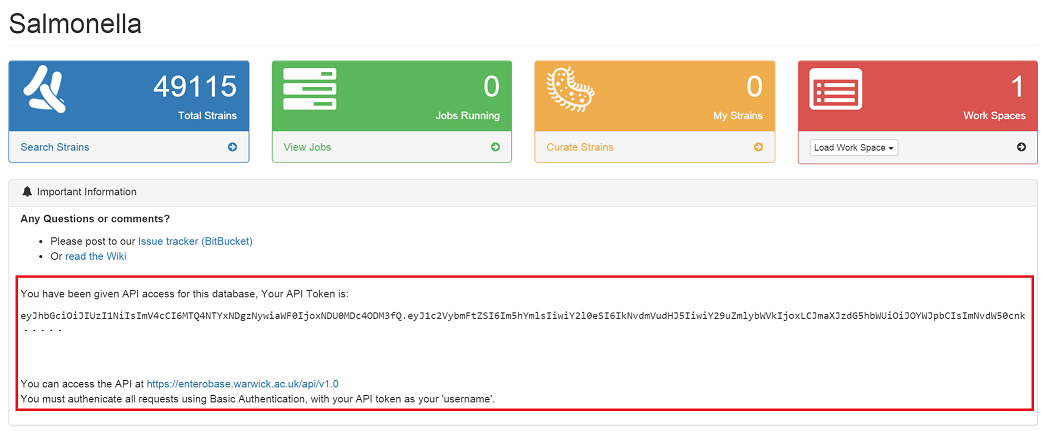
If you DO NOT have API access to a database¶
- Make sure you have an account at https://enterobase.warwick.ac.uk
- Email us at enterobase@warwick.ac.uk with a message something like:
Hi,
I am [YOUR NAME] at [YOUR ORGANISATION]
and I would like access to the EnteroBase Database
[DATABASE OF INTEREST; Salmonella, E. coli, Clostridioides,
Yersinia, Moraxella].
I would like to use the API to [YOUR INTENDED PURPOSE]. This is for a
[COMMERCIAL/ACADEMIC] project about [ONE SENTENCE DESCRIPTION OF YOUR PROJECT].
My username/Email on Enterobase is [YOUR ENTEROBASE USERNAME OR EMAIL YOU USED]
Thanks
[YOUR NAME]
I will get back to you promptly about your request.
Testing out your Token¶
Once you have your Token you can start using it in scripts to download data from EnteroBase. Here is a simple example that just picks out one assembled strain record using curl:
curl --header "Accept: application/json" --user "<YOUR_TOKEN_HERE>:" "https://enterobase.warwick.ac.uk/api/v2.0/senterica/straindata?sortorder=asc&assembly_status=Assembled&limit=1"
You can build requests like this into Python, with a simple example of the same request below:
from urllib.request import urlopen
from urllib.error import HTTPError
import urllib
import base64
import json
API_TOKEN = 'YOUR_TOKEN_HERE'
def __create_request(request_str):
base64string = base64.b64encode('{0}: '.format(API_TOKEN).encode('utf-8'))
headers = {"Authorization": "Basic {0}".format(base64string.decode())}
request = urllib.request.Request(request_str, None, headers)
return request
address = 'https://enterobase.warwick.ac.uk/api/v2.0/senterica/straindata?assembly_status=Assembled&limit=1'
try:
response = urlopen(__create_request(address))
data = json.load(response)
print (json.dumps(data, sort_keys=True, indent=4, separators=(',', ': ')))
except HTTPError as Response_error:
print ('%d %s. <%s>\n Reason: %s' %(Response_error.code,
Response_error.reason,
Response_error.geturl(),
Response_error.read()))
The main important steps to remember are:
- Send your request, usually a GET, with the token added to authorization (Basic) header.
- Data will usually come back in JSON so use a module to cast it to a dictionary.
This is the kind of result you get back. It is usually in JSON, which can be easily treated like a dictionary. Most responses are structured like this:
- links
- paging; links to the previous/next page of data, like webpage pagination.
- Number of records on this page (total_records).
- Total number of records (total_records).
- data, labelled after the endpoint you’ve fetched, in this case ‘straindata’.
- (Data for this record…)
{
"links": {
"paging": {
"next": "https://enterobase.warwick.ac.uk/api/v2.0/senterica/straindata?limit=1&assembly_status=Assembled"
},
"records": 1,
"total_records": 367716
},
"straindata": {
"SAL_FA6876AA": {
"assembly_barcode": "SAL_LA1140AA_AS",
"assembly_status": "Assembled",
"city": null,
"collection_date": 18,
"collection_month": 5,
"collection_time": null,
"collection_year": 2016,
"comment": null,
"continent": "Oceania",
"country": "Australia",
"county": null,
"created": "2016-05-18T09:00:04.867617+00:00",
"download_fasta_link": "https://enterobase.warwick.ac.uk/upload/download?assembly_barcode=SAL_LA1140AA_AS&database=senterica",
"email": null,
"lab_contact": "Vitali Sintchenko",
"lastmodified": "2016-09-06T22:55:53.045179+00:00",
"latitude": null,
"longitude": null,
"n50": 442480,
"orderby": "barcode",
"postcode": null,
"region": "New South Wales",
"secondary_sample_accession": null,
"serotype": "Enteritidis",
"source_details": "NSW ERL",
"source_niche": "Human",
"source_type": "Laboratory",
"strain_barcode": "SAL_FA6876AA",
"strain_name": "NSW29-074",
"top_species": "Salmonella enterica;100.0%",
"uberstrain": "SAL_FA6876AA",
"version": 1
}
}
}
More sample scripts are available at: https://bitbucket.org/enterobase/enterobase-scripts/
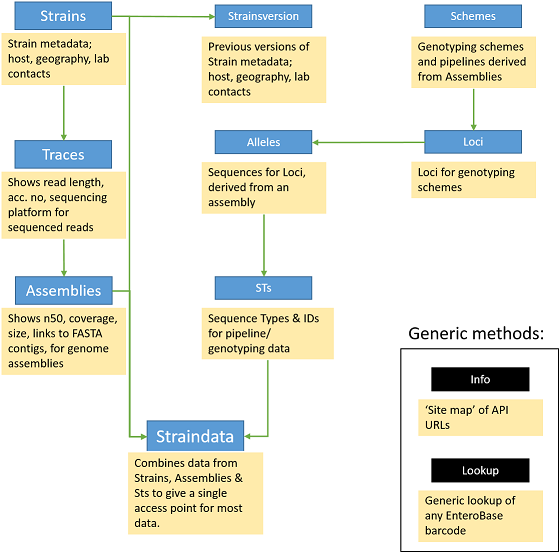
Understanding the EnteroBase API Structure¶
The API tends to follow the logical structure for MLST and NGS data in general. e.g. Strains > Traces > Assemblies, and Loci > Alleles > Sequence types (STs).
There are generic query methods such as Lookup and Info that will help with straightforward lookups of information. If you have any suggestions for new endpoints to help your work, please let us know.
The Swagger sandbox¶
Swagger is an API framework used in the EnteroBase API. It provides interactive documentation of the EnteroBase API, including information about endpoints, inputs, outputs and response codes.
Link to Enterobase’s interactive API documentation (swagger-ui): https://enterobase.warwick.ac.uk/api/v2.0/swagger-ui
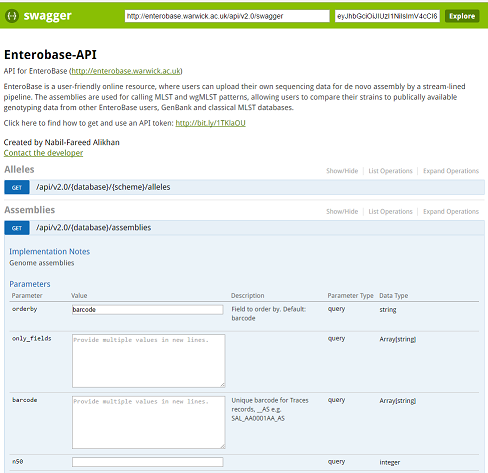
You can just right into playing with requests. There is a demo token already embedded, which is only valid for the Salmonella database (‘senterica’).
Understanding Barcodes¶
Almost all data in EnteroBase is assigned a unique Barcode. This is a unique identifier across all of EnteroBase. It follows a very straightforward structure, split by underscores:
SAL_AA0019AA_ST
- The first part (e.g. SAL) defines the database
- The middle encodes an ID number, letters are used to allow more information per character similar to a UK postcode (CV4 7AL).
- The last part defines the datatype (e.g. ST is Sequence Type record).
Databases are encoded:
| Genus | Tag |
|---|---|
| Salmonella | SAL |
| Escherichia | ESC |
| Yersinia | YER |
| Clostridium | CLO |
| Moraxella | MOR |
Datatypes are encoded:
| Datatype | Barcode tag |
|---|---|
| Schemes | SC |
| Loci | LO |
| Alleles | AL |
| Assemblies | AS |
| STs | ST |
| Traces | TR |
| Strains | None or SS |
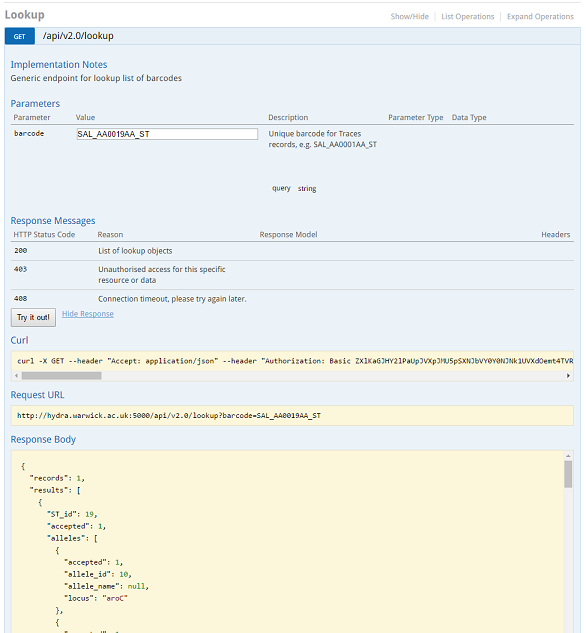
Rapid Barcode lookup¶
Some barcodes can be quickly looked up using the Lookup endpoint in the API. A request is as simple as :
https://enterobase.warwick.ac.uk/api/v2.0/lookup?barcode=SAL_AA0019AA_ST
This gives you a full information on the record, which in this case is about Sequence Type 19 (Salmonella Typhimurium).
{
"records": 1,
"results": [
{
"ST_id": 19,
"accepted": 1,
"alleles": [
{
"accepted": 1,
"allele_id": 10,
"allele_name": null,
"locus": "aroC"
},
{
"accepted": 1,
"allele_id": 7,
"allele_name": null,
"locus": "dnaN"
},
{
"accepted": 1,
"allele_id": 12,
"allele_name": null,
"locus": "hemD"
},
{
"accepted": 1,
"allele_id": 9,
"allele_name": null,
"locus": "hisD"
},
{
"accepted": 1,
"allele_id": 5,
"allele_name": null,
"locus": "purE"
},
{
"accepted": 1,
"allele_id": 9,
"allele_name": null,
"locus": "sucA"
},
{
"accepted": 1,
"allele_id": 2,
"allele_name": null,
"locus": "thrA"
}
],
"barcode": "SAL_AA0019AA_ST",
"create_time": "2015-11-24 19:59:36.295460",
"index_id": 19,
"info": {
"lineage": "",
"predict": {
"serotype": [
[
"Typhimurium",
7372
],
[
"Typhimurium Var. 5-",
440
],
[
"Typhimurium Var. O:5-",
208
]
},
"st_complex": "1",
"subspecies": ""
},
"lastmodified": "2017-02-19 01:44:21.503891",
"lastmodified_by": "admin",
"reference": {
"lab_contact": "DVI",
"refstrain": "9924828",
"source": "mlst.warwick.ac.uk"
},
"scheme": "UoW",
"scheme_index": 1,
"version": 4885
}
]
}
You can play around with this feature in the [interactive documentation](https://enterobase.warwick.ac.uk/api/v2.0/swagger-ui)